Melanoma Mutations
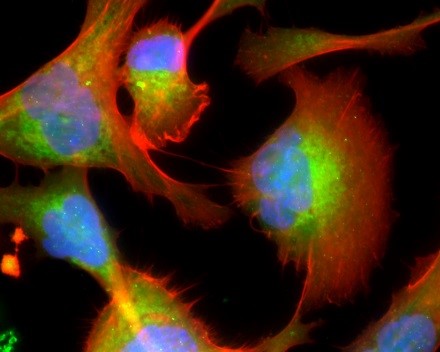
Figure: The melanoma microenvironment. Image courtesy of Dr. Meenard Herlyn, Wistar Institute, Philadelphia, PA.
Malignant melanoma is associated with genetic heterogeneity and a complex etiology. In contrast to other skin cancers, melanoma has a strong tendency to metastasize with a consequently extremely poor overall prognosis for survival. The various phenotypes of melanoma are characterized by clinical features, such as bodily distribution or risk factors. Cutaneous, uveal, acral, and mucosal melanomas have divergent clinical courses and are associated with distinct mutations and risk factors. In the majority of melanomas, a number of genes and signaling pathways are involved in cellular proliferation and growth, and have been implicated in the pathophysiology of melanoma.
We have organized our collection of melanoma cell lines based on the presence and/or absence of mutated or wild-type genes. At a minimum we have characterized these cell lines for the following genes: BRAF, N-RAS, KIT, PTEN and CDK4.
BRAF (BRAFV600E) Mutations
Alterations in signaling cascades that are involved in cellular proliferation and growth have been implicated in the pathophysiology of melanoma. The mitogen-activated protein kinase (MAPK) pathway is a significant driver in melanoma and provides several possible targets for therapeutic intervention. In this pathway, the activation of RAS proteins stimulates the RAF kinases (ARAF, BRAF, and RAF1). The activation of RAF kinases leads to the phosphorylation of the MEK kinases, which in turn phosphorylate the ERK kinases. The RAS-RAF-MEK-ERK signaling pathway that impacts cellular proliferation, differentiation, and survival among many diverse cellular functions, is constitutively activated due to oncogenic mutations in the serine–threonine protein kinase, BRAF. Signaling is initiated when active RAS recruits RAF to the plasma membrane for activation through a complex process requiring lipid and protein binding, conformational changes, and regulatory phosphorylation and dephosphorylation events.
N-RAS Mutations
NRAS-mutant melanoma is a distinct cohort of melanoma that comprises 15% to 20% of all melanomas and appears to confer a poor prognosis. NRAS mutations predominantly in codon 61, lead to increased cellular proliferation and are more potently tumorigenic. NRAS mutations are generally found among melanomas without BRAF mutations, and activating mutations in NRAS confer resistance to BRAF-targeted therapy. Mutations in N-RAS cause activation of downstream serine/threonine kinase MAPK through RAF. N-RAS mutations can also activate other pathways such as PI3K-AKT-MYC, which promote cell cycle progression, cellular transformation, and enhanced cell survival. Although direct inhibition of mutant N-RAS has not been effective, the targeting of downstream pathways such as MAPK pathway inhibitors may prove to be a useful treatment strategy. Furthermore, combined targeting of MEK and PI3K-AKT mammalian target of rapamycin (mTOR) pathways may effectively inhibit N-RAS-mutant melanoma and may provide an alternative therapeutic approach.
KIT Mutations
A small portion of melanomas have changes in a gene called C-KIT. KIT is a type III transmembrane receptor tyrosine kinase. Binding of its ligand, stem cell factor, results in receptor dimerization, autophosphorylation, and activation of several signaling pathways; thereby, mediating cancer cell growth, proliferation, invasion, metastasis, and inhibition of apoptosis. These gene changes are most common in mucosal melanomas derived from the genital regions or mutations in GNA11 or GNAQ genes in uveal melanomas. Some of the targetable mutations in the KIT gene are also found in acral and other mucosal (for example, penile or anal) melanomas but with lesser occurrence. Most KIT mutations are located in exon 11, which codes for the juxtamembrane domain, and in exon 13, which codes for a kinase domain.
PTEN Mutations
PTEN (Phosphatase and tensin homolog deleted from chromosome ten) is a tumor suppressor gene that is mutated in a large fraction of human melanomas. In melanoma, allelic loss or mutations of PTEN have been defined in 5–15% of uncultured melanoma specimens and metastases, as well as in 30–40% of established melanoma cell lines. An ectopic expression of PTEN in PTEN-deficient melanoma cells has been shown to suppress growth, tumorigenicity, and metastasis. The PTEN protein has both lipid phosphatase and protein phosphatase activity. The lipid phosphatase activity of PTEN decreases intracellular phosphatidylinositol (3, 4, 5) -trisphosphate level, ultimately preventing downstream AKT activity. PTEN, containing a phosphatase domain, is inactivated in 12% of melanomas through mutation or methylation.
CDK4 Mutations
The CDK4 is a tumor suppressor gene encoding a protein that helps control cell division. This gene is abnormal in about 5% of individuals with melanoma. CDK4 mutations are relatively common in melanomas in the extremities (such as limbs, fingers, and ears) and mucous membranes (such as those in the mouth, nose, and genitals). Affected individuals possess unique mutations; either an Arg24Cys, located in exon 2 of the gene or Arg24His mutation. The Arg24Cys mutated protein, which was first described as a tumor-specific antigen in a human sporadic melanoma, specifically prevents binding of CDK4 protein to P16A. While there are no FDA-approved drugs that target CDK4, several CDK4 inhibitors are in the early stages of clinical trials.
Melanoma Cell Lines FAQ
How can I ensure that the cells will be viable after resuspension?
Complete adherence to the protocol is necessary to ensure the viability of the cells. Cells should be maintained between 30-95% confluence in tumor-specialized media with 2% FBS (heat inactivated) unless otherwise noted in cell line-specific instructions.
How should I handle the cells before culturing?
Before thawing, frozen cells can be stored in liquid nitrogen or at -80°C only. To thaw the melanoma cells:
- Move the vial from liquid nitrogen to dry ice immediately. Melanoma cells should be thawed quickly. Freeze-thaw cycles can damage/kill the cells and should be avoided.
- Centrifuge the cells at 1500 rpm (500 x g) for 5 minutes at room temperature.
- Discard the supernatant.
- Resuspend the cell pellet in 5 mL of tumor-specialized media with FBS (heat inactivated) and transfer to a T-25 flask.
- Place the flask in a humidified incubator (5% CO2 at 36°C overnight).
- Check the flask after 24 hours for attachment of cells to the flask. If cells are attached, remove the media and add fresh media to the flask.
Which media should I use to grow melanoma cells?
Tumor-specialized media with 2% FBS is the preferred media for melanoma cell lines. However, alternative media (not preferred) recommendations include: 1. DMEM with 5% FBS; 2. RPMI with 5% FBS.
Do you have recommended sources for tumor-specialized media components?
The recommended manufacturers for each component of the tumor specialized media are:
| Product | Source | Item# |
| MCDB-153 | Sigma-Aldrich | M-7403 |
| Leibovitz’s L-15 | Sigma-Aldrich | L1518 |
| FBS (heat inactivated) | Rockland | |
| Calcium Chloride | Sigma-Aldrich | C5670 |
| Sodium Bicarbonate [Added to MCDB-153 preparation] |
Sigma-Aldrich | S5761 |
Please consult the protocol for detailed instructions on the preparation of the media.
What is the optimal growth temperature for melanoma cells?
Melanoma cells grow best at 36°C (not 37°C) with 5% CO2. These incubation conditions must be followed exactly to ensure the viability and growth of the cells.
What is the recommended media for freezing viable melanoma cells?
Freezing media containing 90% FBS and 10% DMSO. Thawed cells should not be frozen again. Melanoma cells should be frozen slowly but thawed quickly.
What is the source of the melanoma cell lines?
The melanoma cell lines were established from patient-derived tumors. Information about the patients or treatments received is not available.
Are melanoma cell lines immortalized?
Melanoma cell lines are considered immortalized because they can replicate indefinitely.